
Transcriptomics with Genomics & Informatics Lab
Transcriptomics focuses on studying the complete set of RNA transcripts expressed in a cell or organism. It provides deep insights into gene expression patterns, helping researchers understand how genes are regulated under different conditions. This powerful approach is essential for identifying biomarkers, disease mechanisms, drug targets, and stress responses in plants, animals, humans, and microorganisms. Transcriptomics is widely applied in biomedical research, agriculture, and biotechnology, making it a cornerstone of modern multi-omics studies.
Analysis Services for Transcriptomics
In the Genomics & Bioinformatics Lab, we offer computational transcriptomics—the all-in data analysis services for RNA-seq projects. Whether studying gene expression, regulatory pathways, or novel transcripts, our services aim to extract the full biological significance of your data.
Transcriptomics enables an understanding of gene regulation, biomarker identification, mechanisms of disease, and responses to stress in plants, animals, humans, and microorganisms. We work with researchers from biomedical, agricultural, environmental, and industrial domains.
🔬 What We OfferWe offer flexible and modular analysis services ranging from very basic expression profiling to advanced transcriptome interpretation. Below are our core offerings:
🧩 Standard Transcriptomics Analyses
1. Quality Control & Preprocessing
✔ Raw sequencing reads clean up to very good quality data
📦 Output: FASTQ QC reports, cleaned data
2. Transcriptome Assembly
✔ Constructs transcript sequences using de novo or reference-based approaches
📦 Output: Assembled transcripts (FASTA), assembly metrics
3. Expression Quantification
✔ Characterizes the abundance of genes and transcripts (TPM, FPKM, raw counts)
📦 Output: Expression matrices, sample-wise summaries
4. Differential Gene Expression (DGE) Analysis
✔ Identifies up/down-regulated genes in conditions
📦 Output: DEG tables, volcano plots, heatmaps, statistics
5. Transcript Annotation
✔ Assigns functions, domains, and homologs to transcripts
📦 Output: GO terms, KEGG IDs, Pfam domains, protein descriptions
6. Alternative Splicing
✔ Identifies transcript isoforms and splicing events
📦 Output: Splicing event lists, isoform quantification, visuals
7. Pathway & Enrichment Analysis
✔ Identifies biological pathways and enriched gene ontology categories
📦 Output: GO enrichment charts, KEGG pathway maps
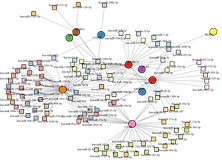
8. Co-expression Network Analysis
✔ Discovers co-regulated gene modules and hub genes
📦 Output: Gene clusters, network graphs, hub gene list
9. Non-Coding RNA Analysis (Optional)
✔ Detects and quantifies miRNAs, lncRNAs, and other regulatory RNAs
📦 Output: Non-coding RNA profiles, predicted targets
10. Proper Data Visualization & Reporting
✔ Provides publication-ready clear graphs and full analysis report
📦 Output: PCA plots, heatmaps, summary charts, project report
🧩Advanced Transcriptomics Analyses
For clients looking for deeper functional insights or systems-level interpretation, we offer advanced transcriptomic solutions:
1. Isoform Switch & Functional Impact
✔ Identifies isoform defining shifts affecting either protein domain or functionality
📦 Tools: IsoformSwitchAnalyzeR
📦 Output: Isoform switch events, predicted functional changes
2. Gene Regulatory Network Inference
✔ Predict interactions between transcription factor-gene and other further regulatory circuits
📦 Tools: GENIE3, GRNBoost, or ARACNe
📦 Output: Network diagrams, TF-target tables
3. Single Cell RNA-seq Data Analysis (if provided)
✔ Clusters single cells and identifies marker genes and infers cell trajectories
📦 Tools: Seurat, Scanpy
📦 Output: Cell-type clusters, UMAP/tSNE plots, pseudotime analysis
4. Time-Series Transcriptomics Analysis
✔ Focuses on how changes in gene expression occurred across time points or stages of development
📦 Tools: maSigPro, DESeq2 time-series modules
📦 Output: Temporal expression profiles, trend plots, key genes
5. Inter-Species Transcriptome Comparison
✔ Compares an expression or annotation profile across species 📦 Output: Ortholog mapping, conserved expression trends
6. Integrated Multi-Omics Data
✔ Integrated transcriptomics, global genomics, proteomics, or metabolomics levels
📦 Output: Cumulative pathway insights, correlation maps, omics overlays.
Gene Network and Pathway Analysis:
-
Construction and analysis of gene co-expression networks.
-
Investigation of spatiotemporal expression patterns.
-
Functional annotation and enrichment analysis.
.jpg)




Single-Cell Transcriptomics:
Single-cell transcriptomics is a molecular technique used to measure the RNA gene expression levels in individual cells, providing detailed insights into cellular diversity, rare cell populations, and developmental processes that are masked in traditional bulk RNA sequencing. It is crucial for understanding cell states, identifying cell types and subtypes, and analyzing cellular heterogeneity in various biological contexts, such as disease progression and tissue development. While powerful, the technique faces challenges in data acquisition and requires advanced computational methods for analysis and interpretation.
Long-Read Transcriptomics:
-
Analysis of long-read sequencing data.
-
Studying full-length transcripts and complex gene structures.
